These are selected publications. For a full list of my publications, see my
Google Scholar profile
.
* denotes equal contribution.
Machine learning
Generative Flows on Discrete State-Spaces: Enabling Multimodal Flows with Applications to Protein Co-Design
International Conference of Machine Learning 2024
Description
We sought to extend flow matching to discrete state spaces. The theory and construction of discrete flows simplifies discrete diffusion which allows for greater flexibility in probability paths, sampling, and target/source couplings. Gat et al. extended our work by scaling it to larger models, experimenting on more domains (coding, images), and developing more extensions. (In retrospect we should’ve called our method discrete flow matching 🥲.)
Next, we combined discrete and Riemannian flow matching into a multimodal diffusion model called Multiflow for jointly generating protein sequence (tokens) and structure (SE(3)). We achieved state-of-the-art results at the time for protein generation. We briefly investigated co-dependency and mutual information properties between the discrete and continuous flows. There is still a lot to understand in multimodal generation.
SE (3) diffusion model with application to protein backbone generation
International Conference of Machine Learning 2023
Description
I wanted to develop a generative model over AlphaFold’s SE(3) protein representation with the goal of enabling structure-based protein design with generative models. I collaborated with the authors of Riemannian score matching (Bortoli et al.) to extend their theory to SE(3) then developed a modified version of AlphaFold’s SE(3)-invariant attention-based neural network for SE(3) diffusion. We introduced a widely used benchmark that measured generation quality, diversity, and novelty to assess protein structure generation. This work was used as the foundation of RFdiffusion (see below) in collaboration with David Baker.
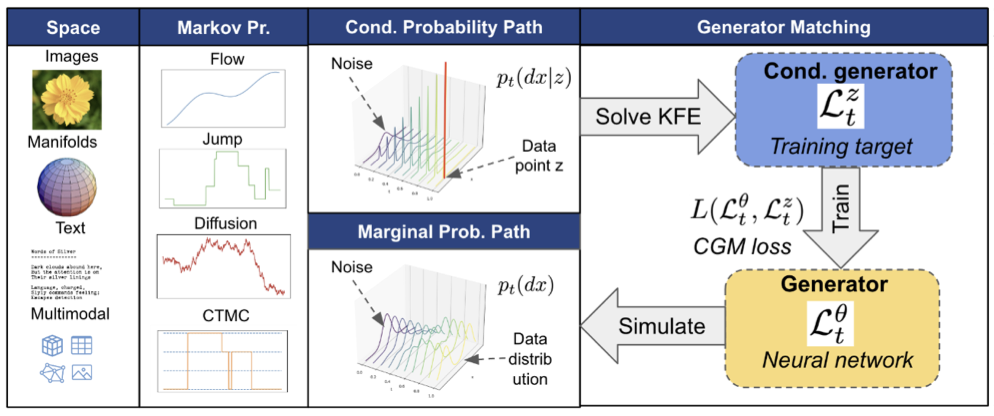
Generator Matching: Generative modeling with arbitrary Markov processes
International Conference on Learning Representations 2025 (Oral)
Description
Generator Matching (GM) is a framework of constructing generative models over Markov processes on arbitrary state spaces. My prior work on Riemannian and discrete flow matching are special cases of GM. I implemented the protein experiments where we demonstrated using GM for the superposition of Markov processes that improved performance in multimodal generation. I like this general framework to encapsulate flows and diffusion on different state spaces.
Improved motif-scaffolding with SE (3) flow matching
Transactions on Machine Learning Research 2024
Description
This work was an continuation of my SE(3) diffusion model where I extended Riemannian flow matching (Chen et al.) to SE(3). As expected, the theory and methodology were much simpler than Riemannian diffusion. My co-authors fixed some numerical instability with exponential and logarithmic maps that greatly improved training stability. Concurrent works (Bose et al., Ajay et al.) demonstrated a low-tempearture sampling trick by scaling the vector field during reverse integration that we also found to improve performance. We applied the model to a protein inpainting task called motif-scaffolding and demonstrated state-of-the-art results. We investigated guided sampling with twised sequential monte carlo (Wu et al.) but found this did not work well without a large number of particles.
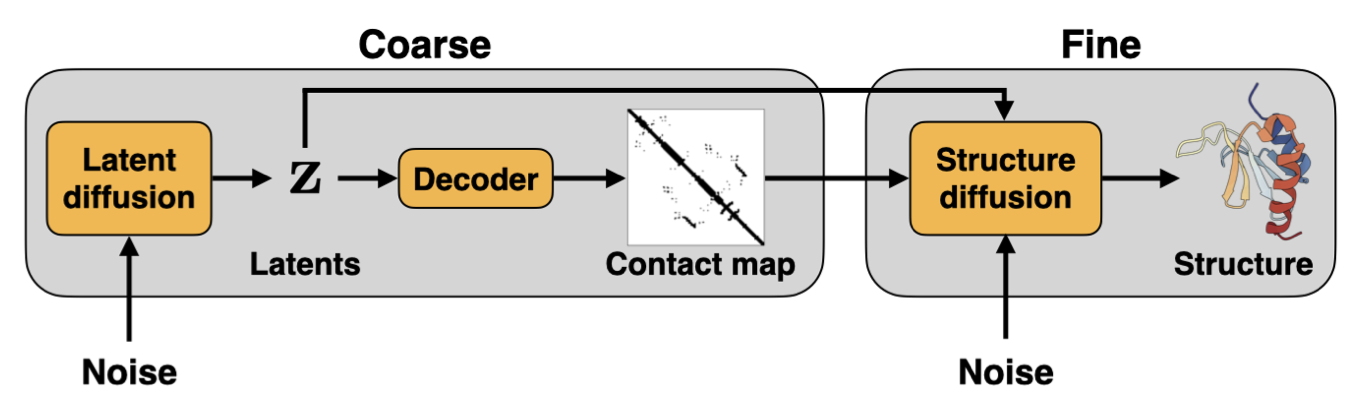
Hierarchical protein backbone generation with latent and structure diffusion
ICLR 2025 Workshop on Generative and Experimental Perspectives for Biomolecular Design
Description
Latent diffusion excels at capturing semantically rich features in the latent space then uses a powerful decoder to generate data conditioned on the latent. We sought to learn a semantically rich latent space of protein topologies then train a hierarchical two-stage diffusion model. First, latent diffusion generates a protein topology. Second, conditioned on the sampled latent, structure diffusion generates the protein structure that adheres to the topology. We found reward optimization to be effective in this framework by doing the guidance in the latent space first then generating diverse protein structures conditioned on the latents. This approach was a proof of concept of how reward guidance could more effective in the latent rather than ambient space. Unfortunately I did not see this project to completion before graduating.
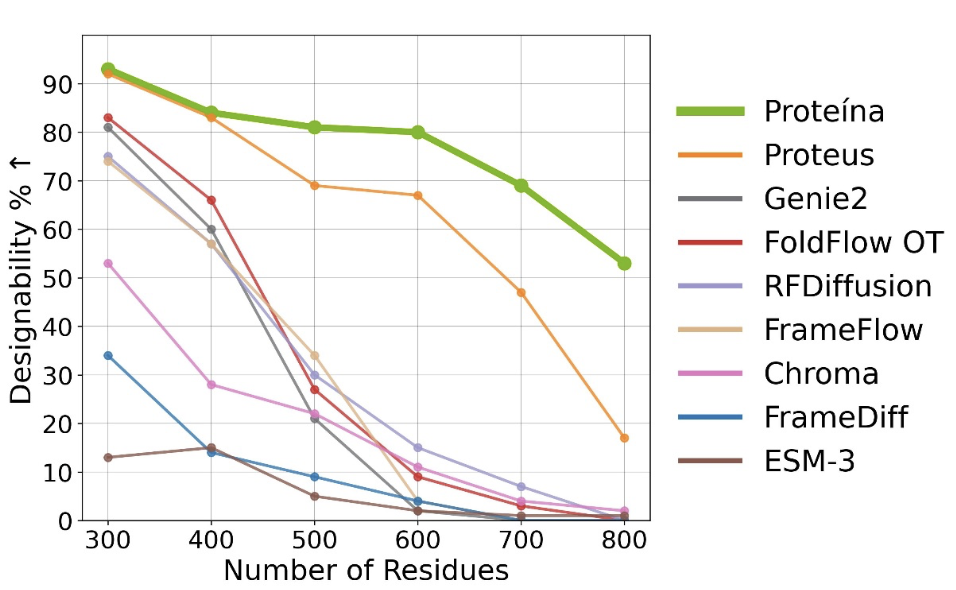
Proteína: Scaling Flow-based Protein Structure Generative Models
International Conference on Learning Representations 2025 (Oral)
Description
I was a advisor providing high-level feedback and guidance. The Nvidia team explored the “bitter lesson” for protein generation by scaling up non-equivariant diffusion models with a Diffusion Transformer architecture up to 400 million parameters and datasets up to 21 million structures. They used classifier-free guidance, autoguidance, and LoRA to guide generation towards desired protein topologies. The model demonstrated state-of-the-art generation quality and diversity while being able to extend to longer proteins up to 800 residues. Other methods often struggle at scaling up to proteins beyond 400 residues (see graph).
Improving protein optimization with smoothed fitness landscapes
International Conference on Learning Representations 2024
Description
We started off wanting to do reinforcement learning (RL) for protein sequence optimization but it turned into a big benchmarking project of 7 different methods after we found serious flaws in the previous benchmarks: sometimes the test examples were 99% similar to examples in train. We constructed a new de-leaked benchmark and evaluated each method. We observed that each method had merits but the real issue was the noisy predictions of the reward model. We explored different regularization strategies and found minimizing the total variation (i.e. increasing the smoothness) (Zhou et al.) could greatly improve test performance. We also found a very simple approach of using Gibbs With Gradients (Grathwohl et al.) with a good reward model would outperform all the more complicated methods using RL, GFlowNets, LLMs, etc. The takeaway? A good reward model is all you need.
Diffusion probabilistic modeling of protein backbones in 3D for the motif-scaffolding problem
International Conference on Learning Representations 2023
Description
This work was the first proof-of-concept of using a diffusion model to generate small, toyish protein structures. The performance was poor but we demonsrated a semantically rich latent space could be learned that smoothly interpolated between different protein structure topologies (see gif). We additionally demonstrated the first application of Sequential Monte Carlo (SMC) to guide the diffusion trajectory towards different topologies.
Science
De novo design of protein structure and function with RFdiffusion
Nature 2023
Description
I collaborated with David Baker and his students to apply my SE(3) diffusion model to protein design. It worked spectacularly where we took a pre-trained structure prediction model and fine-tuned it with a SE(3) diffusion loss. After adding some conditioning capabilities, we tried the model, RFdiffusion, across every protein design task in David’s lab and found it outperformed all prior methods. The AI-designed proteins were then synthesized in the wet lab and found to harbor the functions that we designed them for. This was the first instance of a single generative model successfully designing novel proteins that could bind and catalyze. The fact that a single model could solve multiple tasks and generalize beyond the training set was exciting. There has been widespread adoption by scientists with even a dedicated team to document and improve the software around RFdiffusion.
Scalable emulation of protein equilibrium ensembles with generative deep learning
Science 2025
Description
During my Microsoft internship, I worked with Frank Noe on a protein structure SE(3) diffusion model called BioEmu for sampling protein dynamics. Protein (or molecular) dynamics in simple terms is simulating how proteins move and interact with other molecules to understand their functions. The main idea of BioEmu is to investigate data scaling by first pre-train on publicly available protein structure datasets and then fine-tune on protein dynamics data, both public and in-house generated with large scale compute from Microsoft Azure. The goal is to learn a distribution that samples proportionally to the equilibrium distribution, i.e. sampling proportionally to the amount of time a protein is in a certain pose. BioEmu achieved state-of-the-art performance on capturing the equilibrium distribution of out-of-distribution proteins. If molecular dynamics can scale and generalize to new proteins, then lots of new therapeutic would become possible to develop.
Atom-level enzyme active site scaffolding using RFdiffusion2
Nature Methods 2025
Description
RFdiffusion works for many cases but struggles with enzyme design. Enzymes require atomistic precision to achieve a desired reaction. We extended RFdiffusion with a atomistic representation and my SE(3) flow matching model to achieve atomistic precision when designing enzymes. We developed a novel conditioning approach that separates the design problem into designing the catalytic residues and the scaffold separately but conditioning the diffusion to make sure the catalytic residues and scaffold are compatible. The new model, RFdiffusion2, achieved state-of-the-art results on computational benchmarks for enzyme design and is used inside David Baker’s lab for designing novel enzymes. De novo enzyme design can unlock new chemical reactions not possible through traditional methods such as plastic degradation and new therapeutics.
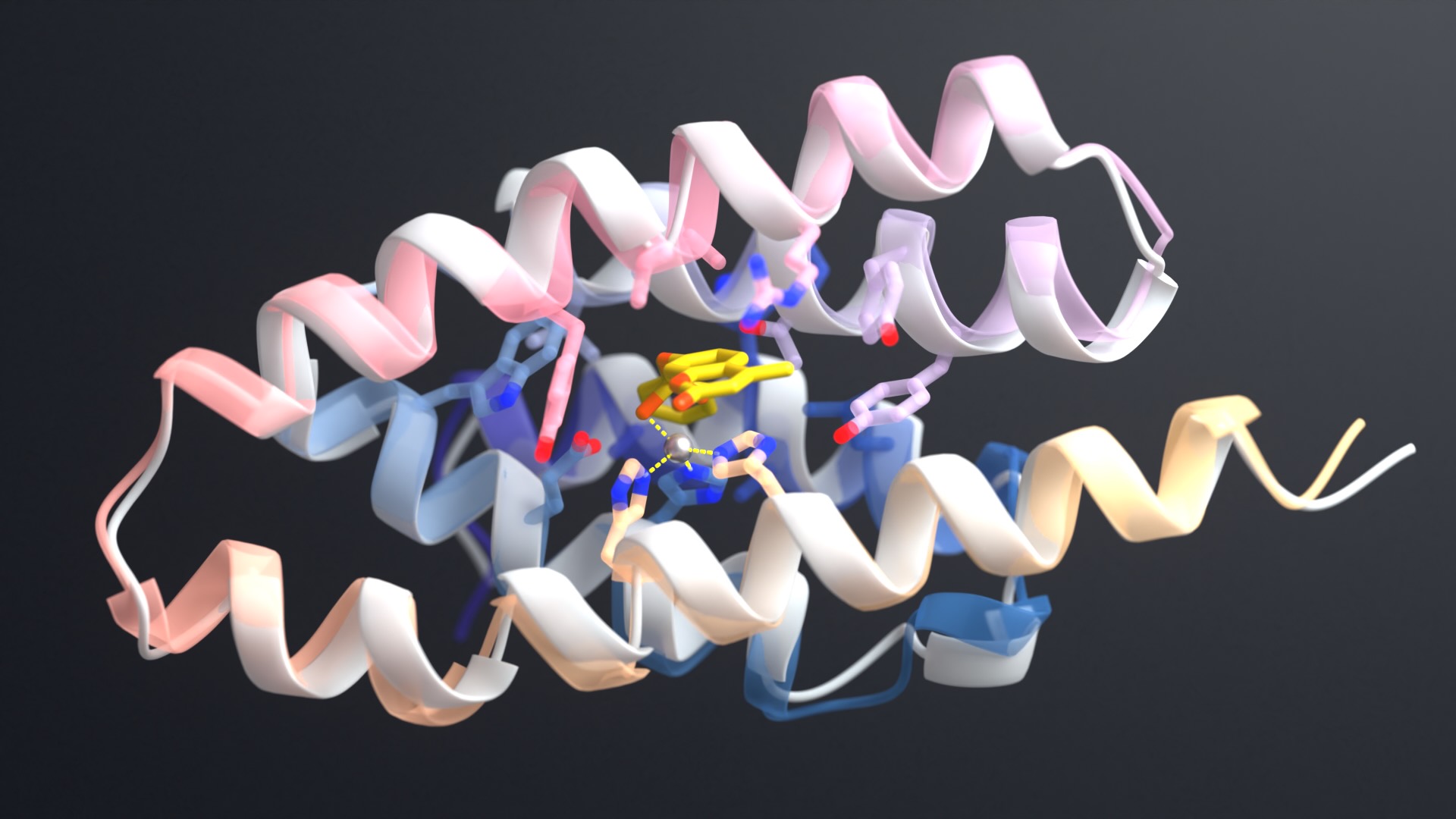
Computational design of metallohydrolases
Nature 2025
Description
Scientists in David Baker’s lab used RFdiffusion2 to computationally design the most active metallohydrolases to date. This is the closest instance of a de novo, i.e. novel, enzyme approaching the same catalytic efficiency as enzymes that nature created through evolution. The biggest hurdle in enzyme design is achieving the same catalytic efficiency as nature and so the results in this work is a big step towards that goal.
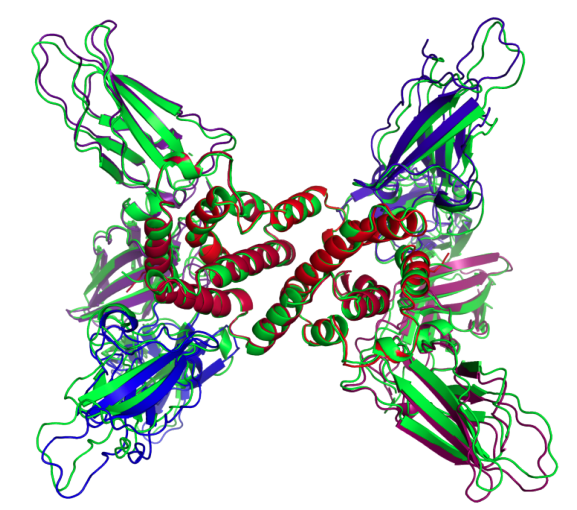
Protein complex prediction with AlphaFold-Multimer
bioRxiv 2022
Description
AlphaFold-multimer (AFm) was the last project I worked on at DeepMind. AFm is the middle child between AlphaFold2 (AF2) and Alphafold3 (AF3). We developed a lot of the data processing, evolutionary sequence modeling, and evaluation that would end up being used in AF3. The main changes in AF3 were the inclusion of arbitrary molecules and the diffusion model that replaced the equivariant graph neural network in AFm. I was a core research engineer working on data processing, evaluation, training, and exploring improvements to the graph neural network architecture for scaling up to large proteins. Unfortunately transformers just scale a lot better!
Predicting conversion to wet age-related macular degeneration using deep learning
Nature Medicine 2020
Description
My first project at DeepMind and first time on a large scale deep learning project. I was the sole research engineer responsible for data, training, evaluation, and collaborating with human doctors. We trained a large 3D U-Net (large at the time) on gigapixel medical volumetric scans to predict if the patient would develop eye disease in the next 6 months. We tried lots of modeling ideas that attempted to incorporate time series of how the 3D scans changed over time (hence modeling 4D data). But the rregular time intervals between scans and small number of scans per patient led to the 4D model not outperforming the 3D U-net. Real world data, especially medical data, is notoriously messy. Data ingestion and cleaning took a significant amount of time. We worked with optometrists to relabel the data and ran a prospective study of comparing the model’s performance against 6 optometrists out of which the model performed better than 5.